Resumen
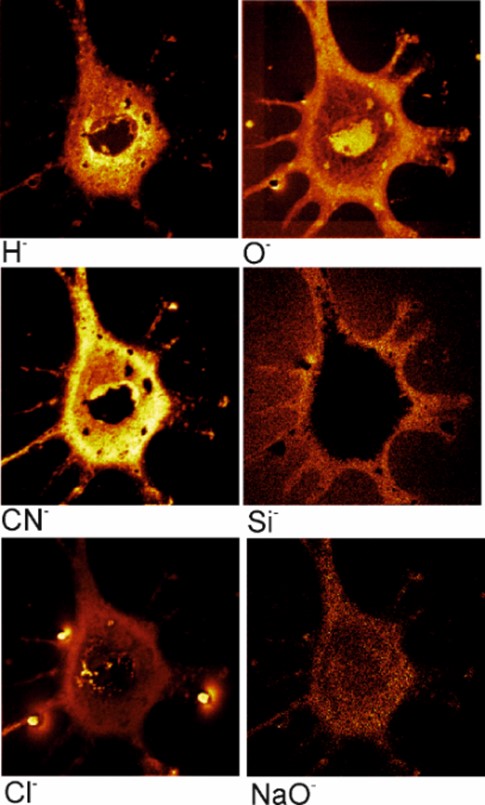
In this paper, we have shown the results of the analysis by the SIMS method of mouse fibroblast cells (3T3-F44A2 line) grown on the silicon surface and fixed by using different chemicals. It was concluded that the standard method of dehydration and fixation of cells with the help of glutaraldehyde allows us to prepare relevant samples for analysis by the SIMS in ultrahigh vacuum. Ion images of chemical elements inside cells with a sufficiently high lateral resolution were obtained, allowing us to study, among other things, the internal structure of cells. The use of SIMS looks preferable for the analysis of cellular metabolism, cell differentiation, ischemia and other processes in cells and tissues grown using special medications and (or) reagents with an artificial isotopic composition of stable isotopes.
Citas
. E.J. Lanni, S.S. Rubakhin, J.V. Sweedler, J. Proteomics 75, 5036 (2012).
https://doi.org/10.1016/j.jprot.2012.03.017
. K. Chughtai, R. M. A. Heeren, Chem. Rev. 110, 3237 (2010).
https://doi.org/10.1021/cr100012c
. J. Vickerman, N. Winograd, Cluster ToF-SIMS imaging and the characterization of biological materials, in Cluster Secondary Ion Mass Spectrometry: Principles and Applications, Ed. C. M. Mahoney (John Wiley & Sons, 2013) p. 269.
https://doi.org/10.1002/9781118589335.ch8
. C.R.M. Grovenor, K.E. Smart, M. Kilburn, B. Shore, J.R. Dilworth, B. Martin, C. Hawes, R.E.M. Rickaby, Appl. Surf. Sci. 252, 6917 (2006).
https://doi.org/10.1016/j.apsusc.2006. 02.180
. J.S. Fletcher, J.C. Vickerman, Anal. Bioanal. Chem. 396, 85 (2010).
https://doi.org/10.1007/s00216-009-2986-3
. P. Agui-Gonzalez, S. Jahne, N.T.N. Phan, J. Anal. At. Spectrom. 34, 1355 (2019).
https://doi.org/10.1039/C9JA00118B
. J. Fletcher, Biointerphases 10, 018902 (2015).
https://doi.org/10.1116/1.4907727
. Y.W. Fana, F.Z. Cuia, L.N. Chen, Y. Zhai, Q.Y. Xu, I-S. Lee, Appl. Surf. Sci. 187, 313 (2002).
https://doi.org/10.1016/S0169-4332(01)01046-7
. A.T. Marshall, P.L. Clode, R. Russell, K. Prince. R. Stern, J. Exp. Biol. 210, 2453 (2007).
https://doi.org/10.1242/jeb.003343
. W. Röhmer, T.D. Wu, P. Duchambon, M. Amessou, D. Carrez, L. Johannes, J.L. Guerquin-Kern, Appl. Surf. Sci. 252, 6925 (2006).
https://doi.org/10.1016/j.apsusc.2006.02.183
. C. Lechene, F. Hillion, G. McMahon, D. Benson, A.M. Kleinfeld, J.P. Kampf, D. Distel, Y. Luyten, J. Bonventre, D. Hentschel, K.M. Park, S. Ito, M. Schwartz, G. Benichou, G. Slodzian, J. Biol. 5, 20 (2006).
https://doi.org/10.1186/jbiol42
. M.L. Steinhauser, C.P. Lechene, Semin. Cell Dev. Biol. 24, 661 (2013).
https://doi.org/10.1016/j.semcdb.2013.05.001
. G. McMahon, C.P. Lechene, Current Protocols 1, e290 (2021).
https://doi.org/10.1002/cpz1.290
. D.J. Wilkinson, Mass. Spectrom. Rev. 37, 57 (2018).
https://doi.org/10.1002/mas.21507
. M.L. Steinhauser, A.P. Bailey, S.E. Senyo, Ch. Guillermier, T.S. Perlstein, A.P. Gould, R.T. Lee, C.P. Lechene, Nature 481, 516 (2012).
https://doi.org/10.1038/nature10734
. C.E. Diaz-Velasquez, F. Castro-Muñozledo, W. Kuri-Harcuch, J. Cell Biochem. 105, 147 (2008).
https://doi.org/10.1002/jcb.21810
. W. Kuri-Harcuch, F. Castro-Muñozledo, Rev. Invest. Clin. 36, 377 (1984).
https://pesquisa.bvsalud.org/portal/resource/pt/lil-32882
. C.R. Ferreira, W.A. Gahl, Transl. Sci. Rare Dis. 2, 101 (2017).
https://doi.org/10.3233/TRD-170015
. R.D. Palmiter, Proc. Natl. Acad. Sci. USA 95, 8428 (1998).

Esta obra está bajo una licencia internacional Creative Commons Atribución 4.0.
Derechos de autor 2023 Array